Reproducible Computational Science on High Performance Computers:
Rachel Slaybaugh
My name is Rachel Slaybaugh and I am an Assistant Professor in the Nuclear Engineering Department at the University of California, Berkeley. I study computational methods for neutron transport: numerical methods for solving the Boltzmann equation applied to neutral particle interactions. The methods I study are both deterministic (e.g. finite difference, etc.) and stochastic (Monte Carlo). I develop these algorithms for reactor design and analysis, radiation shielding, and nuclear nonproliferation applications. Much of my work has an emphasis on high performance computing. (Tagline: intersection of applied math and computational science, informed by nuclear engineering)
Workflow
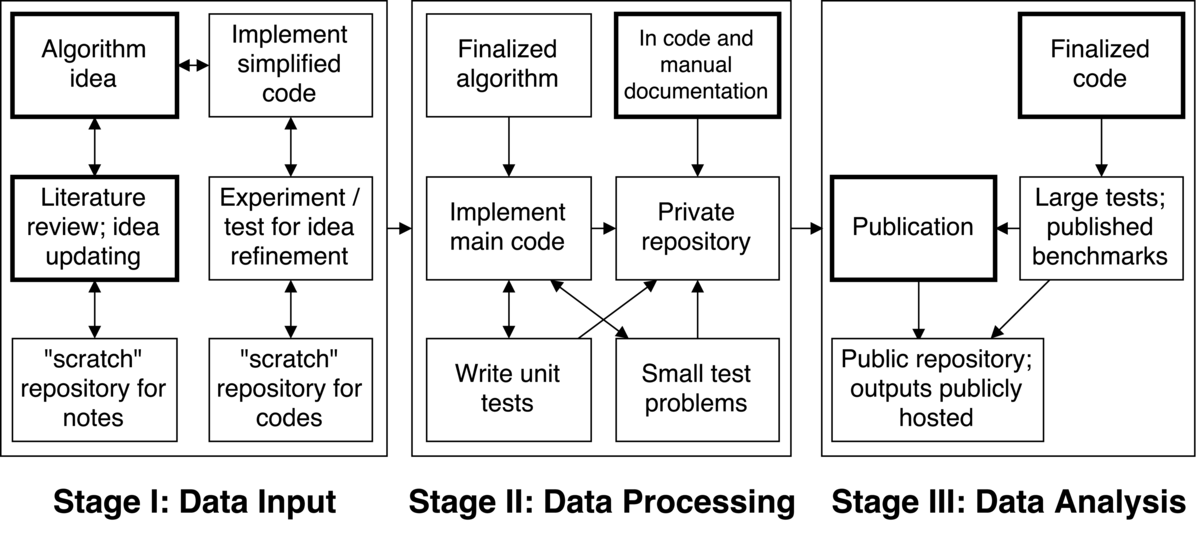 I tend to think of my workflow for code development as having three fundamental steps: (1) idea generation and refinement, (2) idea implementation and testing, and (3) large scale testing and publication.
I tend to think of my workflow for code development as having three fundamental steps: (1) idea generation and refinement, (2) idea implementation and testing, and (3) large scale testing and publication.
Step 1: The starting point of a new project is the development of an algorithm. This comes from a combination of reviewing literature, discussion with colleagues, familiarity with challenges in the field, and so on. As I refine an idea, I find I need to review more literature; as I research the literature, I refine the idea. The algorithm development tends to be collaborative as it is based on discussions with others, but the literature review tends to be independent. I like to write notes while reading papers in one large LaTeX document and keep that document in a repository with all of my other notes and reviews so all of my notes on a given topic are in one place and I only have one place to look for things I have researched in the past.
Next, I implement a simplified version of the algorithm to make sure that it works at all. For example, I would implement a 0D or 1D version (as opposed to 3D) of a method quickly and simply in Python to use for testing. In this step there can be iteration between the algorithm idea and the test code, informed by additional literature review as necessary. Once satisfied with the experiments with the simple code, the algorithm is considered "final" (though of course it can be adjusted later if needed). I am not sure that this part of the workflow is reproducible in the sense that the process could be exactly replicated, but version controlling everything makes it possible to recover intermediate steps, which in some ways allows the idea refinement to be traced.
Step 2: Once there is a finalized algorithm, it gets implemented in the "real" code that has multiple developers and is written in a compiled language like C++. The repository for the code is typically private because, as mentioned, these codes are not completely open. It is often the case that only one or a very few people are working on this idea, so we make a branch and do the development there. I add unit tests to a testing framework associated with the code as I go (for example GoogleTest); the tests reside in the same repository as the main code. As the code approaches completion, I use small "system" test problems to investigate basic system functionality: does the code get the correct answer for analytical/known solutions? what does basic performance look like? etc. The small tests are also version controlled--either in the same repository as the source code or in a separate one.
Once the unit tests are deemed sufficient and, combined with the small tests, everything indicates that code is correct, I finalize documentation. Throughout development I typically use Doxygen to comment the code. Doxygen automatically generates documentation from source code comments when those comments are made using particular, simple annotations. Doxygen works for languages like C++, Python, Java, and others. Using Doxygen is useful for creating an application program interface quickly and easily. However, some extra work is often required to get the theory down and provide clearer directions for using the new algorithm. All of that is written in LaTeX for incorporation into the user and/or theory manual. The documentation LaTeX files are version controlled, often in a separate documentation directory. At this point the code will be reviewed and merged into the main code base. Once the code is finalized, the unit test and small test results should all be reproducible by the other developers--the people who have access to the developer repository.
Step 3: Once there is "finalized" code, it is time to do the real demonstration testing for publication. This involves running large test problems that demonstrate performance of the new algorithm for problems of interest as well as comparison to benchmarks to demonstrate correctness. The literature review, algorithm description, and results of the large (and sometimes small) tests will go in a final LaTeX document for journal submission. Recording of work for journal publication will also be version controlled, typically in a public GitHub repository. The idea is that, beyond the text writeup, the large test input files will be in the same repository as any scripts required to process data and generate plots, all with corresponding directions. Thus if you have access to the code and the repository with tests, scripts, and results, you can rerun all the calculations and process the data.
Pain points
There are a few pain points: An annoying one is getting the documentation right. It seems like just using Doxygen is not enough. To get something that really is user-manual quality you have to write a lot of things twice, just slightly differently. I try to reuse as much as I can, but if things are replicated there is the challenge of maintaining consistency.
A tough one is ensuring that the version of what is released in the end is actually reproducible. This requires the extra step of documenting which version of the code was used (the results should not change in the future, but it is better to have the version clearly written just in case). In principle one can figure this out from the repositories, but if everything isn't stored together that becomes more challenging. Providing directions about how to run everything and which versions of third party libraries were used is also some extra work.
A final pain point is re-implementing the algorithm from the simple case to the complex case, since the simple code is never really used for anything. However, this is a pretty small issue since the toy code didn't take long to develop.
Key benefits
The largest benefit of this approach is having confidence in the validity of the data that you publish. For me that confidence starts with implementing the methods and their tests at the same time. I think everyone should have a unit testing system; it is difficult for me to see how one could have confidence in the correctness of their software without one. I get very nervous about using code that I write if I haven't written tests to go with it.
Having an up-to-date application programming interface is also very useful. When I'm interfacing or working with a piece of code I wrote a long time ago I would not otherwise remember what it did or how to use it. It is also helpful when interfacing with parts of the program other people wrote. This extends to proper documentation. I personally can't remember many things. I must write them down for future reference. Keeping a user and theory manual means not only that users and other developers will know what the code does and why, it means next week I will also know what the code does and why.
I also find that having little bits of experimental code, the low-dimension test pieces I write at the beginning, are useful to have on hand. This does not particularly impact reproducibility, but it is useful to have chunks of code to start with when playing around with new ideas. Similarly, having a repository with literature review notes is good for remembering past research, speeds up writing papers and documentation, and provides a place to start looking the next time.
Key tools
They key tools I use are Doxygen, git (for version control), LaTeX, and plotting and data manipulation tools (usually in Python).
Questions
What does "reproducibility" mean to you?
The first way I think of reproducibility is "can I/my lab reproduce the results in my paper exactly?" After that, "can an independent researcher, given that they have legal access to the required data and software, reproduce the results?" Nuclear engineering data and codes are often controlled, so for many projects only researchers within my field will have access to the required data and software. Fortunately, such non-open-source codes are typically available at no cost to researchers through a simple licensing process.
Why do you think that reproducibility in your domain is important?
The codes that we write are often used to investigate new nuclear systems and make long-term policy or design decisions based on the results. They are also used to study existing nuclear systems. This is important stuff; the codes need to be right and the results need to be verifiable.
How or where did you learn about reproducibility?
Mentors: my PhD advisers valued reproducible practices and insisted that we used them
Student groups: the Hacker Within
Practice: taking on a project that used good practices was how I really learned many of these skills
Community exposure: spending time with others who value reproducible practices
Self-study: looking up things I saw people do that looked helpful
What do you see as the major challenges to doing reproducible research in your domain, and do you have any suggestions?
The biggest challenge is legal: only some people can access the codes and data required. A secondary challenge is access: some of the work I do requires high-performance computing that is not readily available to many.
What do you view as the major incentives for doing reproducible research?
Ethical mandate: I want my work to be right and for others to be able to know that it is right.
Impact: My ideas and products might then be adopted and built upon.
Are there any best practices that you'd recommend for researchers in your field?
Testing, testing, testing.
Would you recommend any specific resources for learning more about reproducibility?
Software Carpentry; the new Scopatz-Huff book.