Achieving Full Replication of our Own Published CFD Results, with Four Different Codes
Olivier Mesnard and Lorena A. Barba
We are members of a computational research group led by Prof. Lorena Barba at the George Washington University in the department of Mechanical and Aerospace Engineering. We do our best to accomplish reproducible research and have for years worked to improve our practices to achieve this goal. According to the "Reproducibility PI Manifesto," pledged by Barba in 2012, all our research code is under version control and open source, our data is open, and we publish open pre-prints of all our publications. For the main results in a paper, we prepare file bundles with input and output data, plotting scripts and figure, and deposit them in the figshare repository. This case study describes what happened when we set out to complete a full replication of published results from our own group, using different Computational Fluid Dynamics (CFD) codes: a new code developed in our group, an open-source code developed by another group, and an open-source CFD library.
Workflow
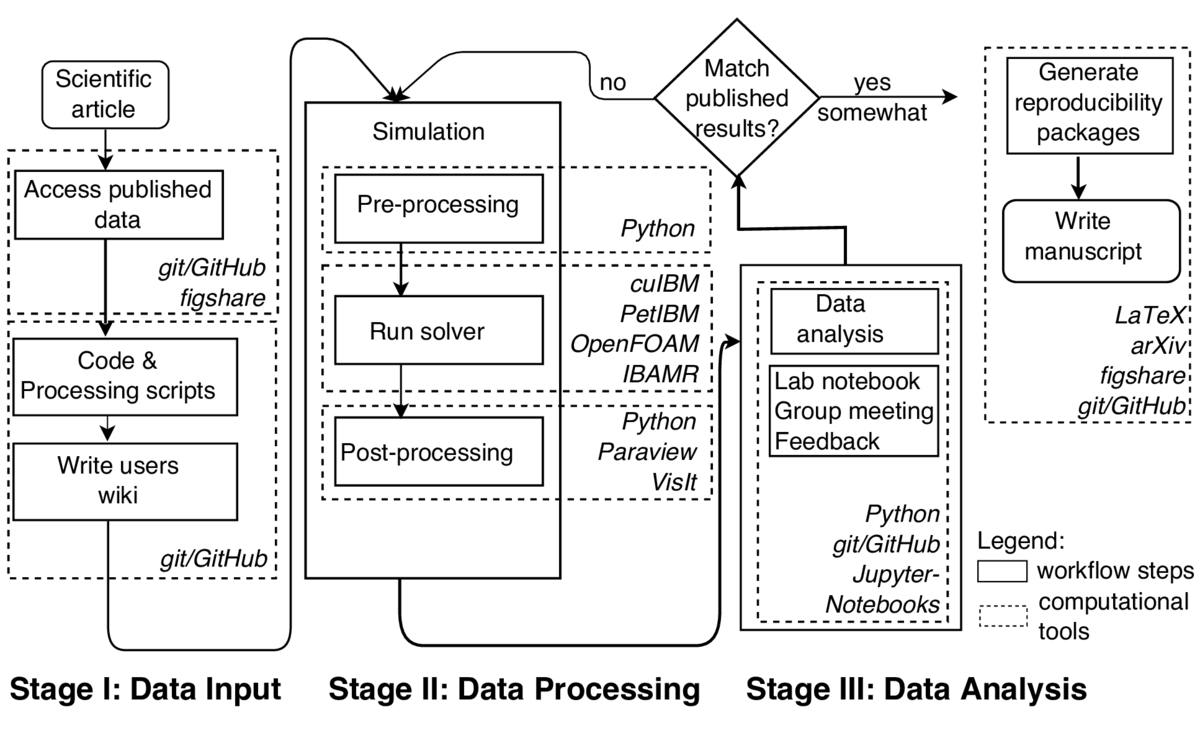 Our research lab has developed over the years a consistent workflow that, we believe, leads to reproducible research. A previous study coming out of our lab, published in Krishnan et al. (2014), already satisfies the criteria of the "Reproducibility PI Manifesto" (Barba, 2012). That work studied the aerodynamics of flying snakes using our code cuIBM for solving the Navier-Stokes equations with an immersed-boundary method. The crux of the study was that, for a particular configuration, the snake's cross-section experiences a lift-enhancement. Here, we describe our effort to achieve full-replication of the main results, using four different Computational Fluid Dynamics (CFD) codes, including cuIBM. We encountered failures and difficulties, leading to improvements in our workflow and conclusions about the challenges for reproducibility in a scenario of highly unsteady flow dominated by vorticity (local spinning of the flow).
Our research lab has developed over the years a consistent workflow that, we believe, leads to reproducible research. A previous study coming out of our lab, published in Krishnan et al. (2014), already satisfies the criteria of the "Reproducibility PI Manifesto" (Barba, 2012). That work studied the aerodynamics of flying snakes using our code cuIBM for solving the Navier-Stokes equations with an immersed-boundary method. The crux of the study was that, for a particular configuration, the snake's cross-section experiences a lift-enhancement. Here, we describe our effort to achieve full-replication of the main results, using four different Computational Fluid Dynamics (CFD) codes, including cuIBM. We encountered failures and difficulties, leading to improvements in our workflow and conclusions about the challenges for reproducibility in a scenario of highly unsteady flow dominated by vorticity (local spinning of the flow).
The first code we used to attempt replication is IcoFOAM: the unsteady laminar solver of the well-known CFD package OpenFOAM. We chose OpenFOAM because it is widely used, open-source, and documented: both code documentation and users' guide are available. With unstructured-mesh finite-volume solvers like IcoFOAM, the mesh generation step is most often what determines the quality of the solution, and we experienced that some meshes resulted in unphysical results. Our first tries led to inconsistent results and we had to replace the mesh-generation tool to get acceptable mesh quality. Setting the boundary condition at the domain outlet was particularly problematic, and made more difficult by lack of documentation for the type of boundary condition we needed. We invested several months of persistent efforts before finally replicating our previous findings (in terms of the lift characteristics) with IcoFOAM.
We then used IBAMR, an open-source library hosted on GitHub that provides several numerical methods for immersed bodies. One of them is specifically designed for non-deforming bodies, which is our situation. Bhalla et al. (2013) published a detailed validation of this method, and some examples are included in the code repository. After many failed attempts, we found that this method requires forcing the fluid to rest everywhere inside the immersed-body, not just at the boundary—this is not an intuitive option with immersed-boundary methods. In the end, we can say that the scientific findings of Krishnan et al. (2014) have been replicated, but we still see noticeable differences in the details of the flow characteristics.
The cuIBM and PetIBM codes are both being developed in our research lab and implement the same immersed-boundary method (Taira & Colonius, 2007). The GitHub code repositories include code documentation with Doxygen, users' documentation (on the GitHub wiki), as well as basic examples and tutorials. cuIBM uses CUSP, an open-source library for sparse linear algebra on a single CUDA-architecture Graphical Processing Unit (GPU). We used cuIBM again to confirm the reproducibility of the published findings in Krishnan et al. (2014). It is important to remark that we had to use the same version of the code, with the same version of the linear-algebra library to obtain the same numeric answers as before. In fact, our first attempts used a newer version of the CUSP library, and failed to replicate the findings! In PetIBM, we use the PETSc library to solve the linear systems on a distributed-memory machine. Even though the mathematical formulation in cuIBM and PetIBM is exactly the same, we observed that a different linear-algebra library could change the results. As of this writing, we have been unable to replicate with PetIBM the lift-enhancement feature of the flying snake.
The lessons learned from this case study are sobering. First, the vigilant practice of reproducible research must go beyond the open sharing of data and code. We now use Python scripts to automate our workflow—all scripts are version-controlled, code-documented and accept command-line arguments (to avoid code modification from users). Instead of using GUIs, we call the Python interpreter included in the visualization tools Paraview and VisIt to plot the numerical solution. Throughout, Jupyter Notebooks and Markdown files document partial project advances. Second, certain application scenarios pose special challenges. Here, we are working with the Navier-Stokes equations applied to highly unsteady flows dominated by vorticity, a particularly tough application for reproducibility. Third, extra care is needed when using external libraries for iterative solution of linear systems: they may introduce uncertainties.
As we now prepare a manuscript to publish the results of this project, it is being written using LaTeX and version-controlled in its own GitHub repository to facilitate collaboration between authors. To advocate open-science, the manuscript will be first available on arXiv. We will also provide, on the repository figshare, a reproducibility package for all simulations and figures reported in the manuscript. These packages include the version of the software, the input parameters, information related to machine architecture, and the necessary scripts to run and post-process the simulation.
Pain points
A critical ingredient in a reproducible workflow is keeping a detailed, up-to-date, and version-controlled lab notebook. It is nearly unthinkable that a proper lab notebook for recording computational experiments could be kept without scripting all steps— pre-processing, running, post-processing—and automaticaly saving command-line inputs. In the project of this case study, we used four different CFD codes in batches of simulations spanning many parameter combinations, resulting in hundreds of runs. The run times varied between 1 and 3 days and the numerical solutions each generated between 3.5 and 16 gigabytes of data. Most of the simulations were run remotely on an HPC cluster at the George Washington University, and the solutions were then moved to several different local desktop machines for post-processing and storage. The lab notebook proved to be vital for tracking all simulations and data. Another aspect of this project that was very time consuming was becoming familiar with new software—it took even longer to familiarize ourselves with codes that offer poor users' documentation. Finally, we also spent considerable time developing automated scripts for analyzing the numerical solutions resulting from different codes (producing different output formats). These scripts, however, are essential to deliver reproducible computational experiments.
Key benefits
In the field of computational fluid dynamics, it can easily take six months or a year to develop software from scratch for solving a specific fluid-flow scenario. On publishing the results, if the authors do not release the code and data used for the study, it leaves any reader hoping to reproduce the results facing a steep time investment. Not surprisingly, studies attempting to reproduce previously published findings are rare. As we have illustrated with our campaign to achieve full replication of our own previous study, there are severe pitfalls and challenges in fluid-flow simulations under unsteady, highly vortical regimes. It is a distinct possibility that many published studies report wrong results. As noted by Leek and Peng (2015), increasing the level of reproducibility of published studies will help uncover flawed research findings. For this reason, the minimum level of reproducibility—making code and data available—is essential for increasing the confidence on any new scientific claims to knowledge generated computationally. Going beyond sharing code and data, full automation and digital recording of experimental campaigns offer the best guarantee of being able to extract scientific value from computational experiments.
Key tools
We use the version-control hosting platform GitHub to support our reproducible workflow. GitHub greatly facilitates collaboration when developing numerical codes and documentation. The platform also allows creating wiki pages for users' documentation. We use GitHub to write manuscripts, to record our group-meetings, and to store teaching materials. We also extensively use Python to automate analysis and post-processing. Progress reports and summaries for discussion in group meetings are best presented using Jupyter notebooks, where textual media is combined with code and visualizations. For a digital record of all steps taken in preparing a simulation and running it, bash scripting is essential. We also use Travis CI for running automated testing of the codes whenever a change is to be merged into the main repository.
Questions
What does "reproducibility" mean to you?
The starting point for our understanding of reproducibility is contained in the pledge "Reproducibility PI Manifesto" (Barba, 2012) which includes these steps:
teaching group members about reproducibility;
maintaining all code and writing under version-control;
carrying out verification and validation and publishing the results;
for main results in a publication, sharing data, plotting scripts, and figures under CC-BY;
uploading preprints to arXiv at the time of submission of a paper;
releasing code no later than the time of submission of a paper;
adding a "Reproducibility" statement to each publication;
keeping an up-to-date web presence.
Some of these items have to do with making our research materials and methods open access and discoverable. The core of this pledge is releasing the code, the data, and the analysis/visualization scripts. Already this can be time consuming and demanding. Yet, we have come to consider these steps the most basic level of reproducible research. On undertaking a full replication study of a previous publication by our research group, we came to realize how much more rigor is required to achieve this, in the context of computational fluid dynamics of unsteady flows. We use the term "full replication" in the sense presented by Peng (2011), that is, completing an independent study using new methods to collect new data, arriving in the end at the same scientific findings. In computational fluid dynamics, full replication of the findings can involve using a different code that implements the same numerical method, or a code that implements a different numerical method altogether but solves the same mathematical model. Because we are solving the Navier-Stokes equations—an unsteady and nonlinear model—certain problem scenarios can present particular challenges to replication.
Why do you think that reproducibility in your domain is important?
In computational science, we use simulations and data analysis as tools for the creation and justification of scientific knowledge. This process of knowledge creation, as in all science, must also produce evidence to justify itself. Reproducibility is a way to provide grounds for trusting the scientific findings obtained computationally. Ensuring that a publication (along with the data used to generate the figures) is reproducible makes it easier for others to corroborate (or reject) a scientific hypothesis. Codes and data used to publish results should be version-controlled and open-source to facilitate reproducibility. Donoho and co-authors (2009) mentioned that we develop codes so that they can be used again by strangers and defined strangers as "anyone who doesn't possess our current short-term memory" (including ourselves in some years). We believe that reproducible research can also prevent scientists from "reinventing the wheel" by having to re-create complete software stacks to build from previously published work.
How or where did you learn about reproducibility?
The group's PI, Prof. Lorena Barba, plays an active role in raising awareness about reproducible research. Incoming students joining our research lab must start by learning the different tools mentioned in the "Reproducibility PI Manifesto". The Software Carpentry Foundation (through workshops and online ressources) also contributes to educate our group members and improve our workflow to achieve reproducible research.
What do you see as the major challenges to doing reproducible research in your domain, and do you have any suggestions?
Reproducible research can be time-consuming, requiring rigorous methods and organization. At various moments during the project, we had to pause and ask ourself if our research was currently reproducible. Often, this was prompted by a conversation or questioning during group meetings. In that sense, a strong collaborative culture in the research group, and beyond in the wider community of the discipline, are vital to instill reproducibility practices in computational researchers. Lack of systematic and widespread educational programs that emphasize reproducible research is a serious obstacle.
What do you view as the major incentives for doing reproducible research?
Making your research more reproducible—e.g., providing reproducibility packages along with the manuscript—is a way of showcasing your skills, a medium for communicating research more transparently, and an invitation to give feedback on your work. If the research community is inclined to put more effort in doing reproducible research, it would prevent scientists from reinventing the wheel by rewriting software in order to build from your work. In the long run, it saves resources to achive scientific knowledge growth, both at the level of a community and within a research group.
Are there any best practices that you'd recommend for researchers in your field?
Again, we insist that automating all the computational workflow and diligently maintaining a lab notebook are fundamental to record your research. We try to avoid GUIs as much as possible and prefer to script everything so that analysis can be automated, reproducible, and recorded. This may be time-consuming but surely beneficial in the longer term of a research project.
Would you recommend any specific resources for learning more about reproducibility?
Barba, L. A. (13 December 2012). "Reproducibility PI Manifesto", 10.6084/m9.figshare.104539. Presentation for a talk given at the ICERM workshop "Reproducibility in Computational and Experimental Mathematics". Published on figshare under CC-BY.
Donoho, D. L., Maleki, A., Rahman, I. U., Shahram, M., & Stodden, V. (2009). Reproducible research in computational harmonic analysis. Computing in Science & Engineering, 11(1), 8-18.
Leek, J. T., & Peng, R. D. (2015). Opinion: Reproducible research can still be wrong: Adopting a prevention approach. Proceedings of the National Academy of Sciences, 112(6), 1645-1646.
Madeyski, L., & Kitchenham, B. A. (2015). Reproducible Research–What, Why and How. Wroclaw University of Technology, PRE W, 8.
Peng, R. D. (2011). Reproducible research in computational science. Science (New York, Ny), 334(6060), 1226.
Sandve, G. K., Nekrutenko, A., Taylor, J., & Hovig, E. (2013). Ten simple rules for reproducible computational research.
Software Testing -- Udacity MOOC.
Stark, P. B. (2015). Science is "show me", not "trust me". Blog post
Vitek, J., & Kalibera, T. (2011, October). Repeatability, reproducibility, and rigor in systems research. In Proceedings of the ninth ACM international conference on Embedded software (pp. 33-38). ACM.
References
Bhalla, A. P. S., Bale, R., Griffith, B. E., & Patankar, N. A. (2013). A unified mathematical framework and an adaptive numerical method for fluid–structure interaction with rigid, deforming, and elastic bodies. Journal of Computational Physics, 250, 446–476.
Krishnan, A., Socha, J. J., Vlachos, P. P., & Barba, L. A. (2014). Lift and wakes of flying snakes. Physics of Fluids, 26(3), 031901.
Taira, K., & Colonius, T. (2007). The immersed boundary method: A projection approach. Journal of Computational Physics, 225(2), 2118–2137.