Feature Extraction and Data Wrangling for Predictive Models of the Brain in Python
Chris Holdgraf
My name is Chris Holdgraf, I am a senior graduate student with the Helen Wills Neuroscience Institute at UC Berkeley. My thesis work involves using predictive models to understand how auditory regions of the brain respond to acoustic features.
I am interested in how experience, learning, and assumptions about the world shape the way that we interact with low level features of sound. This involves a lot of computational work, signal processing, and data cleaning, utilizing a number of packages in the Python scientific ecosystem.
Workflow
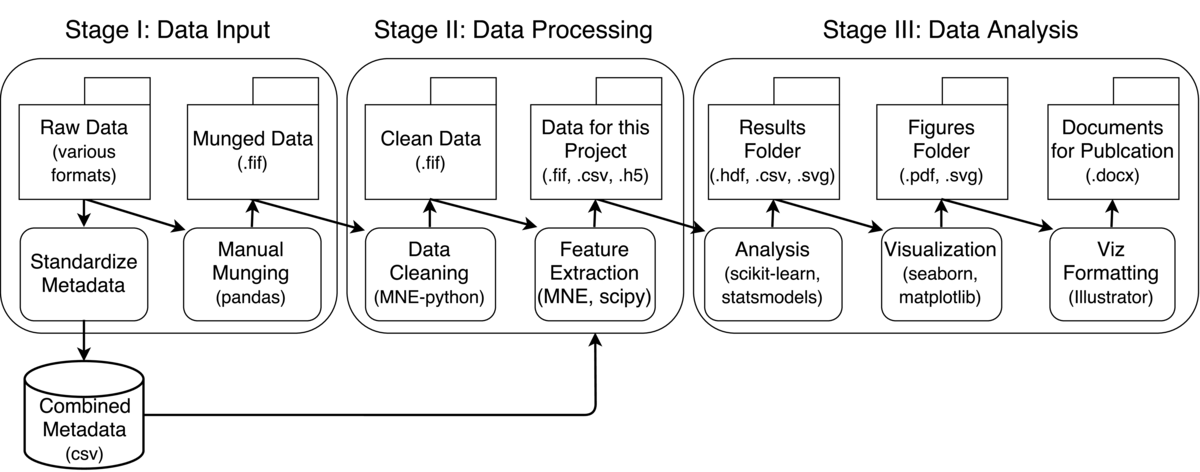 My workflow involves taking raw data from a variety of sources, doing a few steps of munging on each one separately, combining them into a common structured format, and then doing further processing on this data. Here's a general breakdown:
My workflow involves taking raw data from a variety of sources, doing a few steps of munging on each one separately, combining them into a common structured format, and then doing further processing on this data. Here's a general breakdown:
Wrangling raw data
The raw data for my work involves electrophysiology signals collected from the brains of surgical patients in several sites across the country. This is challenging because the nature of the data is quite different from one location to another. I often get the neural data in many different formats, and a loose collection of metadata (e.g., sensor locations, names, sampling rates) that can be anything from a PDF of hand-written notes to a structured text file. As such, the first step in my workflow is to wrangle all of this data into some kind of structured format.
First, I put all data into subject-specific folders. Each of these folders has sub-folders for different kinds of data (e.g., "raw", "munged", "meta"). The sub-folders will eventually be populated with data during processing, and the structure is consistent across all subjects so that I can easily parse them with scripts.
Next, I have a Jupyter notebook that is unique for each person, and is designed to take whatever raw format the data is in and turn it into a standardized version. This is called munge_{subject_name}.ipynb, and will output a file that I can use for the rest of my analyses. Jupyter notebooks are useful here because each subject is different, and will require a different set of steps to get the data ready. For this reason, I like to have lots of plots that go along with the analysis process, and a record of exactly what code was run to create the munged data for that subject.
Because the data often comes in different formats, I make the output of this step the same format for everyone. I use a Python package for neuroscience electrophysiology called MNE-python. This provides a common way of structuring data in order to streamline I/O, processing, and data analysis. I convert all of my raw data into the .fif format, which is a standard format for storing electrophysiology data. This means that I can read the data into other platforms (e.g., R or Matlab) fairly easily. The output files are stored in the folder subject_name/munged.
The final thing I do in this step is look at the metadata files for this subject, double check that all the values inside are correct (this is done with the munging notebook), and then insert them into a "combined" csv file of metadata. I have a Python script called create_combined_meta.py that will look through all the subject folders, find the metadata files that my munging notebook outputs, and turn them all into a single CSV file.
This aggregated CSV has data for all subjects that I have, and makes it much easier to quickly look at information across datasets. To do so, I use Pandas. This is a package that lets you represent tabular-style data in memory, and also gives you "database-style" functionality with their objects (e.g., joining two Pandas objects with partially-overlapping fields). Doing this necessitates that all of my data is in "tidy" format. This simplifies things, because it means that while I have a separate file for each dataset, I have a single combined file across all subjects for their metadata.
I should note that this is the point where somebody usually suggests using a "proper" database like SQL instead of keeping my data in CSV / FIFF formats. I've found that the overhead added by reformatting my data for something like SQL isn't worth the benefit it would give. If I were to start a new project - particularly with larger datasets - then I would likely consider a more robust data storage solution like SQL.
Cleaning the data
Once I've created my munged data, I can now use a single script for processing/analyzing all datasets. In the field of cognitive neuroscience, there are common preprocessing techniques that are carried out in order to improve the quality of the data (e.g., a few filtering steps and rejecting channels that are too noisy). I have a clean_data.py file that will look through the "munged" folder of each subject, load the data, run the cleaning, and then output the results to the folder {subject_name}/clean. This way, I know that any data in the clean folder is ready for further analysis.
At this point, I have cleaned data in each subject's folder, I also have that subject's metadata inserted into a CSV file with all subject information. From now on, I can just load a subject's data file, then load the metadata CSV file for everybody, and query only the rows that belong to the subject that I care about.
Defining a project
When I begin analyzing my data to answer a specific question, I create a new project-specific folder that exists alongside my "data" folder. Each project generally entails a number of analyses, and this is a way to keep them all in the same place. The project folder is structured similarly to the "data" folder. It has a sub-folder for "scripts", for "data", for "results", and for "documents" and any information necessary for publications that come out of this project.
For example, the first thing I might do is create some Python script to extract features of interest. I will put it in project_name/script/feature_name/extract_feature.py. The script assumes that there is data for each subject in the "clean" folder. It will pull the data from "clean," extract whatever feature I'm interested in, and then save the result to the project-specific folder, something like project_name/data/my_feature/{subject_name}_feature.fif. I parse all subject folders and save files in the same manner.
Storing the extracted features for all subjects in a single project-specific data folder makes it much easier to develop scripts/notebooks to further analyze the results, since I don't need to keep track of which features have been extracted for which subjects. I also develop feature extraction scripts using the Sun-Grid engine (a platform for dividing computation between a cluster of computers) for speeding up my analyses. I can do this relatively easily because the folder structure for each subject is the same.
Running analyses
Now that I have a set of features created for each subject, it is time to run analyses and answer questions. These scripts also exist in the project-specific folder, and assume that there is data in the "project_name/data/my_feature/" folder.
A difficulty that I've had is knowing when to keep your analyses in interactive notebooks vs. Python scripts. I generally pilot my analysis interactively - this lets me do sanity checks and on-the-fly calculations that help me develop the final analysis. Once I have code that does a specific analysis, I will put it in a .py script.
The output of this script then goes into a project_name/results/my_analysis folder. They may be in the form of PDFs and SVGs for further inspection, or data files (e.g., CSV) representing model results (such as regression coefficients). For anything consisting of lists, numpy arrays, or simple dataframes, I use the h5io package, which provides a fast way to read/write collections of data to hdf5 files (another standardized data storage format).
Finally, I use the outputs of the analysis script to create visualizations and decide whether or not my analysis actually worked. I use another set of notebooks to read in the results, perform last-second wrangling, statistics, etc, and output visualizations. If I have a publication-ready figure in mind, I will create a notebook specifically for that figure in another folder called project_name/fig_{analysis_name}.
When creating actual figures for papers, I like to use software like Adobe Illustrator to make sure that my fonts are the proper size, well-spaced, etc. I use visualization notebooks to create high-res PNG versions of my data that have most formatting stripped away (except for the data). These plots are then linked to an Illustrator file, so that they are updated automatically when a new plot is created. This way I can easily arrange my plots and standardize fonts without doing a lot of manual tweaking.
Finally, I use Microsoft Word to write drafts which I put in the "doc" folder. These pull from the figures I've created in the "fig" folder. Ideally I would use text files here with LaTeX, but the team that I work with makes this prohibitive.
A general note
This process has been refined many times over the past year, and the original structure looked very different than this. My original file system had code and data living in totally different places. Moreover, it had project-specific scripts and more general utility scripts living in the same place. The goal of this file structure is to keep data and scripts together when they have a natural pairing, and to separate out my more production-ready functions/modules from "hackier" project-specific scripts. Below is a list of some things that I've learned along the way, and that have guided the development of this system:
For any data/code that are project-specific, keep these together in the same general file hierarchy.
Rather than creating metadata structures that live next to the data, come up with a “master” metadata template, then store entries from every subject in a CSV file that follows this template. Rather than storing data in separate subject files, include an entry with “subject id” in the metadata file so that you can pull out individual subjects in this way.
Utilize other packages whenever possible, particularly with the annoying task of data I/O. In my case, I store all my raw data as ‘.fif’ files, which can be opened easily in Python or Matlab with well-supported third party packages. I also use
pandasandh5ioto read / write metadata files, which makes it very easy to slice and dice these files for particular entries that I want.More generally, take a “database” approach to how you store any data. Even though I’m not storing data in a MySQL database per-se, I can still draw inspiration for how this data is organized. Treat data entries as rows, and data features as columns, and then combine / split up the data using pandas database-style syntax (e.g., joins and merges). A great guideline for this is the "tidy" data specification described by Hadley Wickham.
Put ad-hoc code in a project-specific folder. Be much pickier about code that you expose in public Python modules for any project. If you think a function is worth generalizing, then move it out of the project folder and to its proper module, and document it extensively.
Do all coding with automatic PEP8 and PEP257 checkers. PEP8 is a set of standards for naming conventions, code syntax, using white space, etc to ensure that code is clean and readable. PEP257 is a set of similar guidelines for docstrings. Many "fully-featured" text editors have plugins that automatically check code using these guidelines and highlight errors, which is useful for quickly writing clean code.
Make a conscious effort to structure Python scripts differently from Jupyter notebooks. Structure code (and data) such that it lends itself well to scripting, rather than assuming interactive sessions for everything.
Use Jupyter notebooks to glance at the data and preliminary results, but move code into scripts as it becomes more refined or complex. This avoids creating a mega notebook with tons of different analyses in it.
Structure code so that some scripts live with the data that they operate on. E.g., if you’ve got a script that only operates on a specific collection of data, renames specific columns in that data, and always saves it to another location relative to the original data, then create another folder “script” right next to that data folder. Put all data-specific scripts into this folder. This way, you know that scripts operate with relevant data nearby.
Finally, this is not specific to this project but has been very useful to me. Find opportunities to contribute to other open-source projects. Open pull requests and learn about how to use the code. The back-and-forths and input you get will make you a much better coder, and your codebase / research will greatly benefit from it.
Pain points
The hardest part of my workflow has been deciding how to balance flexibility and control. On the one hand, you don't want your scripts to be so specific to data that they break as soon as anything changes, but on the other hand creating code that generalizes well and isn't terribly confusing is really difficult to do. In my case, I initially made errors on both of these fronts, but the current structure of my data seems to be more intuitive, easy to maintain, and easy to grow.
Another big issue I've had lies in dealing with different formats of data and information. For example, I want to version control all of the code that I write, but:
Does that mean that I should create one big repository for all of the code described above? What about a separate repository for each project?
How should I split up the general modules and functions vs. the code that only lives with a particular project?
Finally, how should I deal with the fact that there are lots of other "non-code" files living with this file structure (e.g., images)? Should they be version-controlled, or should the code just assume that the data lives in particular folders? What would happen if somebody else copied the code and didn't get the data?
I don't necessarily have good answers for these issues, and I'm still coming up with a solution that makes me happy, but these are some things that I'm thinking about.
Key benefits
The biggest benefit of this system is the fact that messy, subject-specific data is quickly turned into a standardized format that is consistent across all of my subjects. This is useful because it means you can write scripts that analyze the entire dataset without doing a lot of extra customization. Moreover, because the structure of the filesystem is the same for everybody (including things like naming conventions), it is easy to find what you want from each dataset.
Another benefit is the fact that I am storing code along with the data that it operates on. Some people feel strongly that this is a bad idea, but I've found it to be useful so long as the within-project folder structure still makes sense. In previous workflows, I had all of my code in one folder, and all of my data in another folder. This often led to confusions where I was unsure which code operated on what data. It also made it more difficult to connect the steps of preprocessing and feature extraction chains. Now, if I want to know all of the things that have been done to a collection of data files, I just need to look into its corresponding "script" folder.
Finally, by separating out operations that are true for all projects (e.g., data munging and cleaning) and those that are project-specific, the scope of individual projects becomes more clear and easy to follow. I think of the data pipeline as a single tree trunk, where projects branch out from this trunk and do extra things to the data, on top of the base workflow of preprocessing. Now, my file structure more naturally follows this concept.
Key tools
The two most useful tools that I have found are Pandas and MNE-python. Pandas made it much easier to embed metadata with the signals that I analyze. It allowed me to store information from lots of subjects in a single CSV file, and treat it as a "database" by using queries on it. MNE-python is a package for electrophysiology in neuroscience written in Python. When I discovered it, I found that it duplicated many of the functions I had already written, and in general did this much better than I had. Moreover, it has a lot of convenience functions for doing I/O, which up until then was a pain to maintain. By using these two packages, I was able to significantly cut down on the amount of custom-written functions that I used to wrangle my data.
Questions
What does "reproducibility" mean to you?
The discussion in this writeup covers the first 6 months of the project. To that extent, my definition of "reproducibility" means actually being able to reproduce my own results (aka, coding for my future self). I ran into a lot of issues to keep things streamlined and understandable in my own head, which made it difficult to interpret my findings. Obviously this would generalize to other scientists trying to reproduce my analyses and work as well.
Why do you think that reproducibility in your domain is important?
Because it's a guiding principle that will make my code more understandable, maintainable, and extendable for others and for myself.
How or where did you learn about reproducibility?
At first it came from teaching a few Software Carpentry classes and reading things online. Lately, I have gotten a lot of help by contributing to the MNE-python project, as I've found that going through the pull request process for a well-maintained project is a great way to learn a lot about coding well.
What do you see as the major challenges to doing reproducible research in your domain, and do you have any suggestions?
In my case, legality is a big problem because I'm dealing with medical data that cannot be shared publicly. Another big problem is simply a matter of training and incentive. Right now there is little opportunity to learn how to code well or how to make reproducible science. To make matters worse, I see very little incentive for anyone to actually do so (if they want to be a tenured faculty).
What do you view as the major incentives for doing reproducible research?
Other than the warm fuzzy feeling, I think the biggest advantage is that when you code and organize for other people, you also code and organize for yourself in the future. This makes your life much easier in the long run.
Are there any best practices that you'd recommend for researchers in your field?
Front-load a lot of thinking/planning before you just start creating scripts and functions. Spend a good chunk of time thinking "big picture" early on, then zoom in and build some stuff, then zoom back out and decide if it was a good idea or not. Don't get lost in the weeds.
Would you recommend any specific resources for learning more about reproducibility?
Software Carpentry is a great one, but most other stuff is just scattered on stack overflow unfortunately. I think that finding a good package that has a sweet-spot of contributors (aka, not so few that you don't get feedback, not so many that it's a huge pain to do anything). Try to contribute something via a pull request and learn from the other people in the community. It will be a great way to learn good coding principles. Finally, find a community around you (at the university, at local companies/hackathons, etc) that shares your interests. Spend time learning and teaching with these people.