Turning Simulations of Quantum Many-Body Systems into a Provenance-Rich Publication
Jan Gukelberger and Matthias Troyer
My name is Jan Gukelberger, I am a computational condensed-matter physicist, who recently completed his PhD at the Institute for Theoretical Physics, ETH Zurich. This case study describes a project I worked on during my first year as a PhD student (2010-2011). This case study was conducted together with my advisor, Matthias Troyer.
Broadly speaking, the project's goal was to characterize a family of quantum many-body model systems. A specific model is described by a large matrix, the Hamiltonian, and its physical properties can be deduced from the lowest-lying eigenvalues (energies) and the corresponding eigenvectors (quantum states). Therefore I wrote a C++ program that would build the Hamiltonian matrix for a given set of model parameters, run an iterative diagonalization algorithm, and output the corresponding properties. Analysis of the results produced by this program for different parameters yielded a deeper understanding of the studied model family and corroborated analytical results obtained by colleagues. The analytical and numerical results were finally published together in Phys. Rev. B 85, 045414 (2012).
Workflow
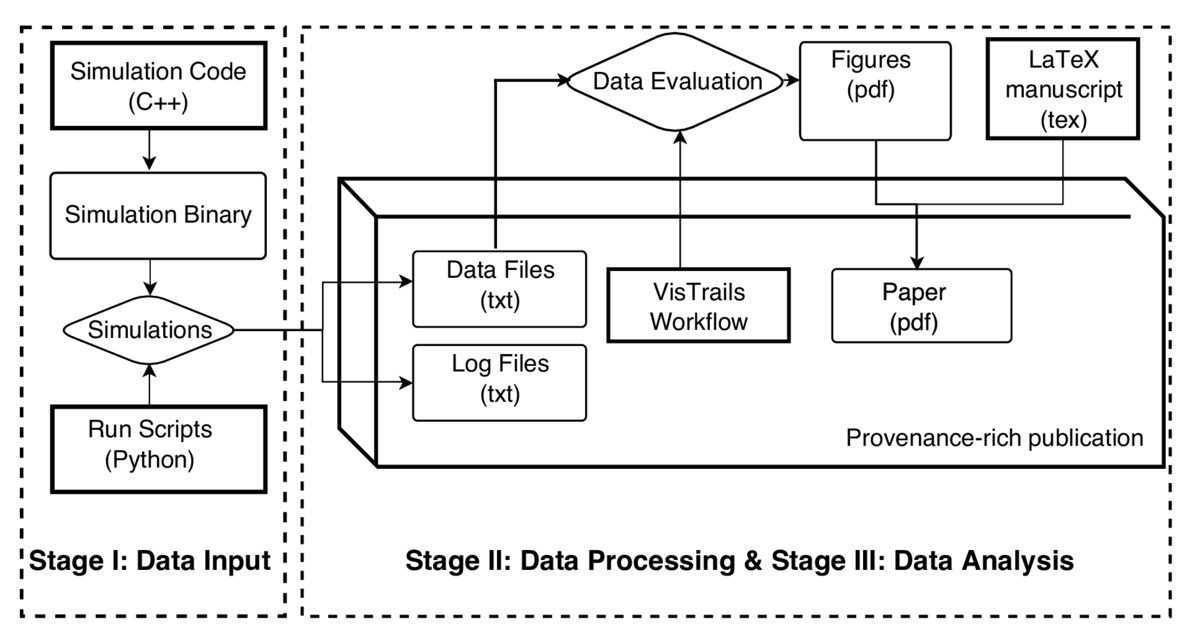 Since the simulations may require a large amount of compute resources (on clusters or large workstations), it is usually not feasible or desirable to re-run the whole process in one go. We therefore typically adopt a two-step approach: The output of the simulation runs is treated as primary/raw data, which is archived along with log files containing detailed information about source code version, execution environment, and input parameters. The evaluation and transformation of this raw data to the final results (typically figures with plots) should then be as easily repeatable as possible, ideally with a single push of a button or script execution.
Since the simulations may require a large amount of compute resources (on clusters or large workstations), it is usually not feasible or desirable to re-run the whole process in one go. We therefore typically adopt a two-step approach: The output of the simulation runs is treated as primary/raw data, which is archived along with log files containing detailed information about source code version, execution environment, and input parameters. The evaluation and transformation of this raw data to the final results (typically figures with plots) should then be as easily repeatable as possible, ideally with a single push of a button or script execution.
In this study, we opted to publish the raw data as supplementary information on the publisher's (APS) web server and provide workflow files for the VisTrails system, which would retrieve the raw data from the server and recreate the figures contained in the paper. VisTrails is an open-source scientific workflow and provenance management system which was used for the data evaluation and plotting tasks in the project. This way, any reader can inspect in detail and rerun all steps of our data analysis.
At the beginning of the project is the development of a simulation code in C++. Once the code is ready, it is used to explore the properties of the physical model under study. For this purpose, it is compiled and run with different input parameters on different systems (workstations and clusters). Because the simulation code is adapted and expanded continuously over the course of the study, it is essential to record what version of the code produced which results. To this end, we use a run script (Python), which records the code version (subversion revision), input parameters for the run, as well as details about the build configuration (compiler, libraries, etc.) and the system environment in which the code is run (host, date & time, dynamic libraries, etc.). All these details are written to a log file next to the data file that contains the results of the simulation. Both are semantically linked to each other by having the same file name, up to the extension (.dat and .log).
These output files constitute the raw data, which is collected on a desktop system for evaluation. The evaluation typically loads data files from several simulations (corresponding to different input parameters), computes some numerical transformations of the data, and finally produces one or more figures (pdf files) with data plots. We code the evaluation process in VisTrails workflows, making use of the ALPS package (delivered with VisTrails), which contains many utility routines for common processes like data transformations, fitting and plotting. We generally aim for a separate workflow (VT file) for each figure. This increases modularity and makes the development of the workflows easier, but implies that several VT files need to be opened and executed if all figures are to be recreated.
Finally, the manuscript of the paper is written in LaTeX, including the figure files created by the VT workflows. LaTeX compilation produces the paper as pdf, which constitutes the central part of the publication.
Publishing the paper together with the raw data and workflows, such that readers could easily inspect and reproduce our data evaluation process, turned out to be a challenge in its own right and required intense interaction with the publisher. Here, the main problem was the need for cross-references between the manuscript, the VT workflows, and the raw data, because the final location of each component only becomes available in the last step of the production process, when the files cannot be changed anymore without manual intervention from the production team. Some aspects of this issue are explained in detail in our report Publishing provenance-rich scientific papers, Procs. TAPP'11. In the end, the publisher was not able to insert links from the figures in the paper to the corresponding workflow files, but only a general reference to the supplementary material section on their server, where all the workflows could be downloaded.
Note: One collaborator actually recreated the figures with a different plotting tool before publication in order to improve their visual appearance. For this purpose, we amended the VT workflows to export the preprocessed data to an external file before plotting. Therefore, the figures presented in the paper are equivalent, but not identical, to the ones created by the VT workflows.
Pain points
Apart from the non-trivial publishing process, the main pain points during the study were connected to the fact that the data evaluation had to be done in the VisTrails GUI and to the opaque-ish VisTrails workflow file format:
Data evaluation (execution of VT workflows) could not be scripted at that time.
The evaluation could not be run on a cluster/via ssh.
Version management was harder because viewing differences between versions was not as easy as looking at the diff file for a Python script.
This also made the synchronization of workflows between different machines (e.g. laptop and workstation) less straightforward.
When now inspecting the "reproducible publication" on the APS server, three years after publication, some mid- to long-term issues become obvious, because both the used software and the publisher's infrastructure is evolving. Continuous testing and maintenance of the published instructions and workflows would be needed in order to keep up with the changes:
The instructions we provided in the supplementary materials section accompanying the article do not work out of the box with the current VisTrails version: In the most recent stable VisTrails release at the time of writing (2.2), the ALPS package is broken and needs to be patched with the latest (not-yet-released) ALPS version. Otherwise initialization of the ALPS package fails and the workflow cannot be executed.
The APS journals were not able to guarantee a long-term stable location for supplementary material. In fact, the URL has already changed, such that the workflows fail to fetch the raw data from the APS server, unless the URL is fixed manually in each workflow. For one specific example, the original location http://prb.aps.org/epaps/PRB/v85/i4/e045414/dyl_ladder_gap.zip has been changed to http://journals.aps.org/prb/supplemental/10.1103/PhysRevB.85.045414/dyl_ladder_gap.zip. Also, the cause of the error is not easy to fix for the uninitiated, because the DownloadFile module actually succeeds (it downloads the html file shown at the old URL), but the subsequent UnzipDirectory module fails with the message "BadZipFile: File is not a zip file". Hence we, the authors, need to prepare new workflows with adapted URLs and send them to the publisher for replacing the original ones whenever their infrastructure changes.
For these reasons I would now prefer to publish a self-contained archive containing the raw data and a script with minimal dependencies in a wide-spread language, such as Python, which reruns the analysis and reproduces the figures. This would be more robust with respect to changes in the publisher's infrastructure. Also, backwards compatibility issues might be expected to be solvable more easily in the long run for scripts in a wide-spread language, compared to special purpose solutions like VisTrails/ALPS (no matter how professional and helpful the developers of the software may be at the moment).
Key benefits
One of the most important points is recording exactly what version of the simulation code was run with what kind of input parameters. This excludes some of the worst cases of "non-reproducible results" and should definitely be a standard practice. (I cannot judge how established this practice is in our field because code and log files are rarely published.)
A second point is the actual publishing of raw data and evaluation workflows, allowing any reader to directly inspect all details of the evaluation process -- even those that the authors did not deem important enough (or forgot) to mention in the paper. This is clearly not widespread practice in our field and would be quite desirable in my opinion.
Questions
What does "reproducibility" mean to you?
In general, given a publication (in a refereed journal), source codes and raw data (which might be available publicly or in the institute's repositories), an expert from my field should be able to understand, and in principle repeat, every step of the study from the running of the correct version of the simulation code to the final results presented in the published paper.
How or where did you learn about reproducibility?
Some basic principles are quite evident, but integrating them in an efficient workflow may require some programming/version control experience. I came into contact with the VisTrails software due to a collaboration between our group and the VisTrails developers, aimed at integrating the evaluation tools of the ALPS package (developed within our group) with VisTrails.
What do you see as the major challenges to doing reproducible research in your domain, and do you have any suggestions?
The main challenge is probably making the recording of provenance data as simple as possible, so no-one has an excuse not to do it.
Another point is that simulation codes, raw data, and evaluation tools are rarely published. Most researchers are very reluctant to publish their codes, e.g. because they do not want competitors to publish results produced with their code before they can, or because they feel ashamed of the poor quality of their code. Raw data may be large and in non-standard format. And the evaluation may be performed by a chain of different tools, which makes publishing of the workflow hard.
What do you view as the major incentives for doing reproducible research?
Apart from research ethics and institutional requirements demanding this, the recording of provenance information can make a researcher's life significantly easier when he/she discovers a discrepancy between different sets of results produced during a single study or in studies by different researchers. (Are the discrepancies caused by differences in the code, different input parameters, or data evaluation?)